Research Review: Specific host metabolite and gut microbiome alterations
- Martian to be
- Jul 25, 2023
- 4 min read
Research Review: Specific host metabolite and gut microbiome alterations are associated with bone loss during spaceflight
🌐Link to the paper: https://www.sciencedirect.com/science/article/pii/S2211124723003108?via%3Dihub=
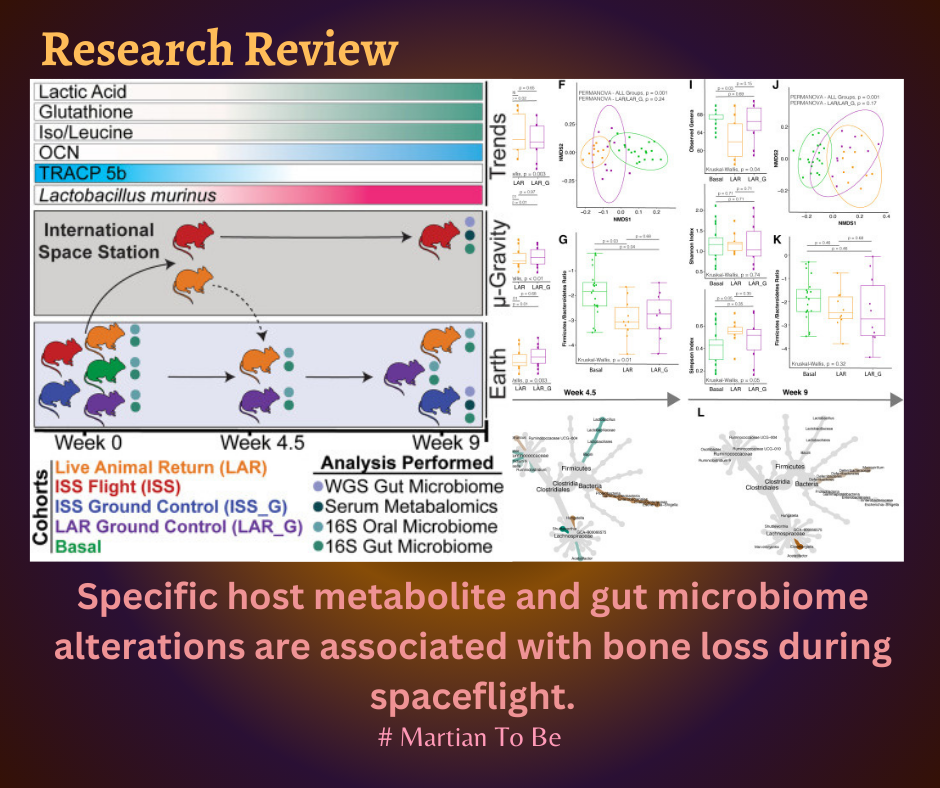
The study investigates the association between specific host metabolite and gut microbiome alterations with bone loss during spaceflight. The study found that the abundance of L. murinus and Dorea sp. is elevated under microgravity exposure, and identified pathways capable of producing lactic acid, leucine, and glutathione are enriched, which are elevated in the serum of flight cohorts, leading to bone loss.
The paper provides new insight into the axis of the murine gut microbiome and bone homeostasis interactions as well as a glimpse into understanding microbiome diversity upon return to Earth in a rodent model. The study also tests the use of a non-ovariectomy BALB/c rodent model as a potential in vivo system for evaluating the effects of microgravity on the host's microbiome and bone homeostasis. The ongoing studies, such as the RR-7 mission and the NASA Astronaut Microbiome project, aim to further clarify the effects of microgravity on the different microbiomes of rodents and humans.
The practical implications of this paper are significant for space exploration and human health. The findings suggest that specific host metabolites and gut microbiome alterations are associated with bone loss during spaceflight. This information can be used to develop countermeasures to prevent bone loss in astronauts during long-duration space missions. Additionally, the study provides insight into the effects of microgravity on the gut microbiome, which could have implications for human health on Earth. The study also tests the use of a non-ovariectomy BALB/c rodent model as a potential in vivo system for evaluating the effects of microgravity on the host's microbiome and bone homeostasis. The ongoing studies, such as the RR-7 mission and the NASA Astronaut Microbiome project, aim to further clarify the effects of microgravity on the different microbiomes of rodents and humans.
The introduction of this paper discusses the adverse effects of low-Earth-orbit space travel on mammalian physiology, particularly the loss of bone mineral density. The paper focuses on the microbiome shifts that occur in microgravity and their potential health consequences. Prior to the Rodent Research (RR) mission series, animal research in microgravity was limited to less than 3 weeks of spaceflight onboard space shuttles with limited sample size. The RR-5 mission enabled live animal return (LAR) of rodents from the International Space Station and investigated the intersection of the gut and oral microbiome ecology and the bone homeostasis axis. The paper also discusses the link between changes in the gut microbiome and bone homeostasis through immune system-modulating effectors, vitamin and nutrient deficiencies, endocrine regulation, and energy metabolism through short-chain fatty acids (SCFAs).
🟣The methods used in this paper are as follows:
🔸Sequencing of the 16S ribosomal V4 RNA gene was performed to analyze the fecal microbiome samples.
🔸Library preparation for sequencing the V4 region of the 16S rRNA gene consisted of amplification and barcoding using the standard Illumina 515f/806r primer set.
🔸After library construction, 2x150bp (paired-end) sequencing was performed on an Illumina HiSeq 2500 platform.
🔸The reads of amplicons from the V4 region of 16S rRNA were processed using the DADA2 package following a standard workflow of quality trimming, de-replicating, DADA2 denoising, read-pair merging, and chimera removal steps.
🔸A total of 454 distinct amplicon sequence variants (ASVs) were identified by DADA2 among all samples.
🔸Fecal ASV sequences were searched against a reference sequence set containing 16S rRNA gene sequences from all named prokaryotes downloaded from the SILVA high-quality ribosomal RNA database(v132) using ''blastn''.
🔸The best hit covering R 95% of the query length was identified for each sequence.
🔸The genus level read count data, generated by the "tax_glom" function of Phyloseq were used in this analysis.
Note: This paper focuses on the microbiome shifts that occur in microgravity and their potential health consequences. The methods used in this paper are related to the analysis of fecal microbiome samples.
The data used in this paper is related to the analysis of fecal microbiome samples from mice that were exposed to microgravity during NASA's RR-5 mission. The authors used sequencing of the 16S ribosomal V4 RNA gene to analyze the fecal microbiome samples. After library construction, 2x150bp (paired-end) sequencing was performed on an Illumina HiSeq 2500 platform. The reads of amplicons from the V4 region of 16S rRNA were processed using the DADA2 package following a standard workflow of quality trimming, de-replicating, DADA2 denoising, read-pair merging, and chimera removal steps. A total of 454 distinct amplicon sequence variants (ASVs) were identified by DADA2 among all samples. The genus level read count data, generated by the "tax_glom" function of Phyloseq were used in this analysis.
The results of the paper suggest that specific host metabolite and gut microbiome alterations are associated with bone loss during spaceflight. The study found that the abundance of L. murinus and Dorea sp. is elevated under microgravity exposure. The authors also identified pathways capable of producing lactic acid, leucine, and glutathione that are enriched in the gut microbiome of mice exposed to microgravity.
These metabolites were found to be elevated in the serum of flight cohorts, which lose bone homeostasis. The study also found that the gut microbiome of mice exposed to microgravity had an altered F/B ratio, a metric for gut dysbiosis in GI disease states. Overall, the study suggests that microgravity exposure can lead to changes in the gut microbiome that may have negative health consequences, such as bone loss.
We value your ideas and feel free to comment below.💜



Comments